Code
library(dplyr)
library(tidyr)
library(ggplot2)
library(mice)
library(forcats)
library(ggExtra)Imanol Zubizarreta
February 25, 2024
Missing data is one of the most important issues to consider when it comes to data/statistical analysis. Different methodologies have been proposed to deal with it and significant efforts on validating/evaluating them have been made. The starting step of these evaluations is usually simulating data accompanied by the missingness addition/simulation under different scenarios and different missing proportions. The action of adding missingness in your data set is called amputation and this very first step will be the main topic in this post. The scenario under which you will be generating missing data will be certainly the most important step in your simulations, thereby worth creating an amputation specific post. You will see, I am avoiding any clinical trial specific verbatim 🙂, as I want these starting posts to be domain agnostic.
The primary objectives in this post are:
The secondary objectives are:
In the clinical trial setting, which is the main topic on my daily work, you may have longitudinal data, and you may certainly be willing to estimate the difference in the outcome between different treatment groups in an specific time point. However, for the seek of focusing on what we want to emphasize in this post, we will simulate very simple analysis data sets, as all the terms we will be talking about in this post are not clinical trial domain specific. These are the three variables we will be simulating for this post.
SUBJID AGE OUT
1 SUBJ-1 34.39524 93.85686
2 SUBJ-2 37.69823 94.82552
3 SUBJ-3 55.58708 130.31676
4 SUBJ-4 40.70508 128.78221
5 SUBJ-5 41.29288 98.07033
6 SUBJ-6 57.15065 164.63071Different missing proportions in the outcome variable will be simulated now based on the complete data set generated above, as these data sets will potentially be used in some later posts, where some evaluations will be made as a function of missing proportion.
In this post, and with the aim of keeping things simple, the outcome (OUT) variable will be the only variable at risk of facing missingness. The AGE variable will be always complete.
Before we go into the amputation process, worth briefly presenting the three scenarios under which you can be generating missing data. We do not need complex/longitudinal data sets to understand these concepts:
Let’s describe the main parameters of the mice::ampute function.
SUBJID AGE OUT
[1,] 1 1 0 SUBJID AGE OUT
[1,] 0 1 0As commented before, we will be generating missingness under MAR, where, long story short, the older subjects will have a higher hazard of being missing as we are stating the parameter type = RIGHT, under a continuous probability distribution (we will later talk a bit more about it). You can either use the odds parameter if you want to use a quantile based likelihood of being missed by assigning different relative probabilities to each age quantile (do not forget to set the cont parameter to FALSE).
Let’s make use of the function now! Please make use of Rianne Margaretha Schouten et al for more details on the arguments of mice::ampute.
#calling mice::ampute under different target missing proportions
amputed_list <- lapply(missing_rates, FUN = function(missing_tmp) {
set.seed(123)
result <- mice::ampute(
anl, prop = missing_tmp,
patterns = pattern_missing,
mech = "MAR",
weights = weights,
type = "RIGHT")
})
#showing the first records of the amputed data set with 60% of missingness
amputed_list[[3]]$amp[1:10, ] SUBJID AGE OUT
1 <NA> 34.39524 NA
2 <NA> 37.69823 NA
3 <NA> 55.58708 NA
4 <NA> 40.70508 NA
5 <NA> 41.29288 NA
6 <NA> 57.15065 NA
7 <NA> 44.60916 NA
8 <NA> 27.34939 NA
9 <NA> 33.13147 NA
10 <NA> 35.54338 95.40559Ups! mice::ampute() is converting the character variable SUBJID to NA. Let’s add the subject variable information again in the four amputed data sets (under the four different missing proportions).
SUBJID AGE OUT
1 SUBJ-1 34.39524 NA
2 SUBJ-2 37.69823 NA
3 SUBJ-3 55.58708 NA
4 SUBJ-4 40.70508 NA
5 SUBJ-5 41.29288 NA
6 SUBJ-6 57.15065 NA
7 SUBJ-7 44.60916 NA
8 SUBJ-8 27.34939 NA
9 SUBJ-9 33.13147 NA
10 SUBJ-10 35.54338 95.40559Let me check whether the amputation has been done under MAR assumption. Always good to check it out!
breaks <- quantile(amp_df_list[[1]]$AGE, c(.33, .67, 1))
breaks <- c(0, breaks)
labels <- c('young', 'medium', 'old')
amp_df_bind <- do.call(rbind, amp_df_list) %>%
bind_cols(
miss_prop = rep(names(amp_df_list), each = nrow(amp_df_list[[1]]))
) %>%
mutate(is_missing = ifelse(is.na(OUT), "Missing", "Not Missing"),
AGE_CAT = cut(AGE, breaks = breaks, labels = labels))
ggplot2::ggplot(amp_df_bind , aes(x = AGE_CAT, fill = is_missing)) +
geom_bar() +
facet_wrap(~ miss_prop) +
theme_bw()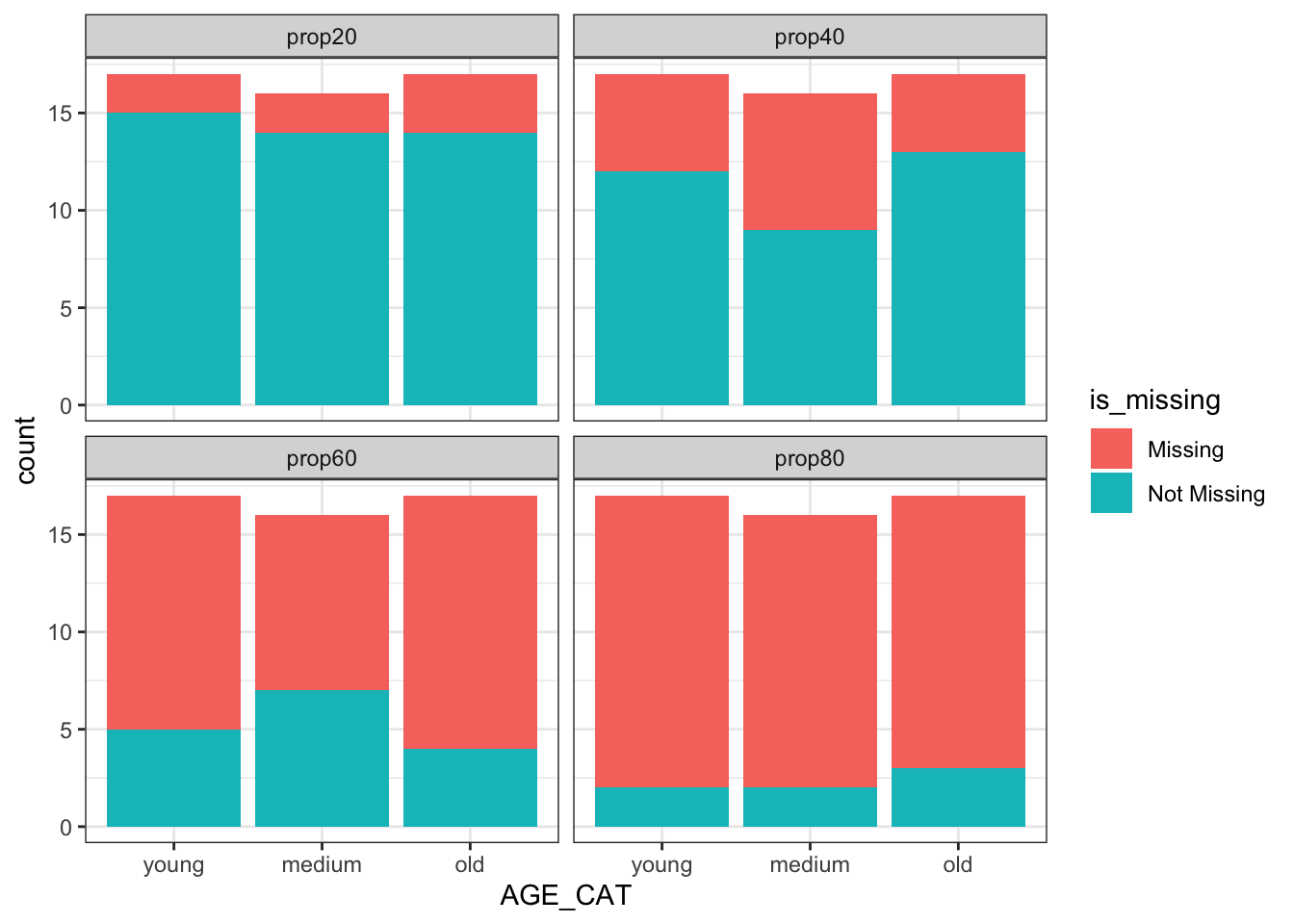
Not what we were expecting, right? Missingness in OUT variable is by no means dependant on the age variable 🤔, which was our original intent!
After digging into the inner functions from mice::ampute, I realized the user must remove the character variables such as SUBJID before going over the amputation process. Please see the issue I created in case you are interested in the details. Once said this, keep this in mind when you will be simulating missingness in your analysis data sets, as you may not be getting the intended missing distributions.
After going over the same process but removing the SUBJID variable, you can see the distributions make much more sense. Missingness does depend on age now. 🥇
AGE OUT SUBJID
1 34.39524 93.85686 SUBJ-1
2 37.69823 NA SUBJ-2
3 55.58708 NA SUBJ-3
4 40.70508 NA SUBJ-4
5 41.29288 NA SUBJ-5
6 57.15065 NA SUBJ-6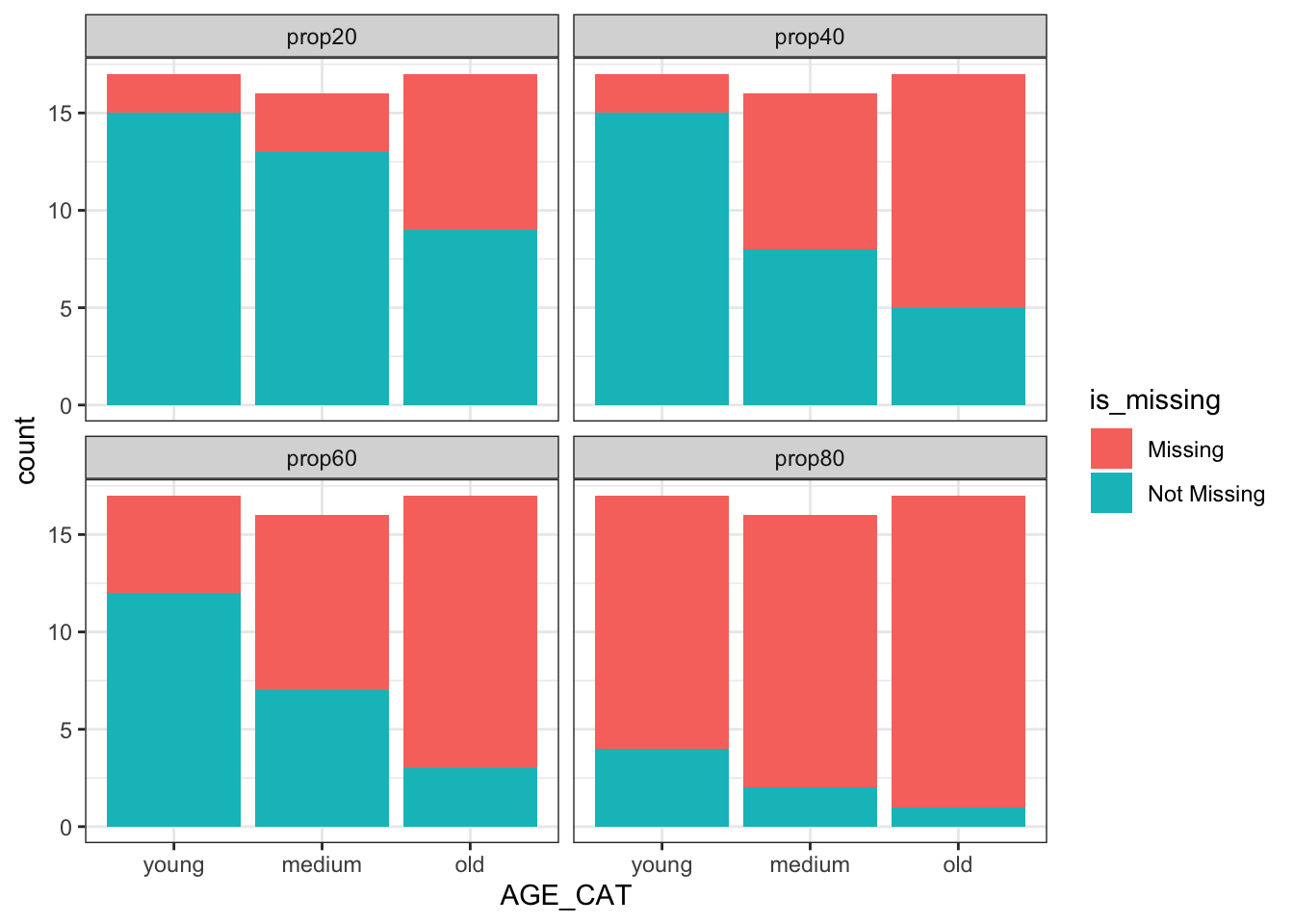
I would strongly recommend you going over Schouten et al. (1) (take your time on Figure 2) to understand how mice::ampute() works when there are more than one missing patterns (multivariate amputations). You will need to pay attention to freq parameter of the function.
Let’s mimic some of the steps mice::ampute is carrying forward.
Worth mentioning the weighted sum scores which is defined by the following equation.
\[ wss_i = w_1*y_{1i} + w_2*y_{2i} + .... w_m*y_{mi} \]
Where:
As we have seen when defining the weight parameter under mice::ampute, we have only assigned a non-null weight (of value 1) to the variable AGE, as we were aiming to ampute the OUT variable under MAR assumption based on how old the subject is. The above equation would be very simple in this example (\(wss_i = w * AGE\)), where w = 1.
Last but not least, I would like to briefly talk and visualize the continuous probability distribution (logistic distribution) mice::ampute is generating to decide on the missingness. First we get the scaled weighted sum of scores.
[1] -0.64250835 -0.28576479 1.64634862 0.03899559 0.10248111 1.81522403By using the above scaled scores, we will be describing the probability of being missing by a logistic distribution function.
See in the following plot the pattern among the values of the weighted sum scores and associated probabilities of being missing. I color-coded by their original categorical AGE variable so that you can see the older subjects will be at higher risk of being amputed by the use of logistic distribution function. Empty points are the ones that mice::ampute converted them to missing in the above missing generation steps (under missing proportion rate of 40%).
\[P(missing) = \frac{e^{x}}{1 + e^{x}}\]
#using plogis to get the missing probabilities from exp(x)/(1 + exp(x))
p_missing <- plogis(scores)
p_missing_scores <- data.frame("p_missing" = p_missing, "scaled_wss" = scores) %>%
bind_cols(AGE_CAT = amp_df_bind[51:100, c("AGE_CAT", "is_missing")])
ggplot(
p_missing_scores,
aes(x = scores, y = p_missing, fill = AGE_CAT, color = AGE_CAT, shape = is_missing)
) +
geom_point(size = 3) +
scale_shape_manual(values = c(1, 19)) +
scale_size_manual(values = c(2)) +
theme_bw() 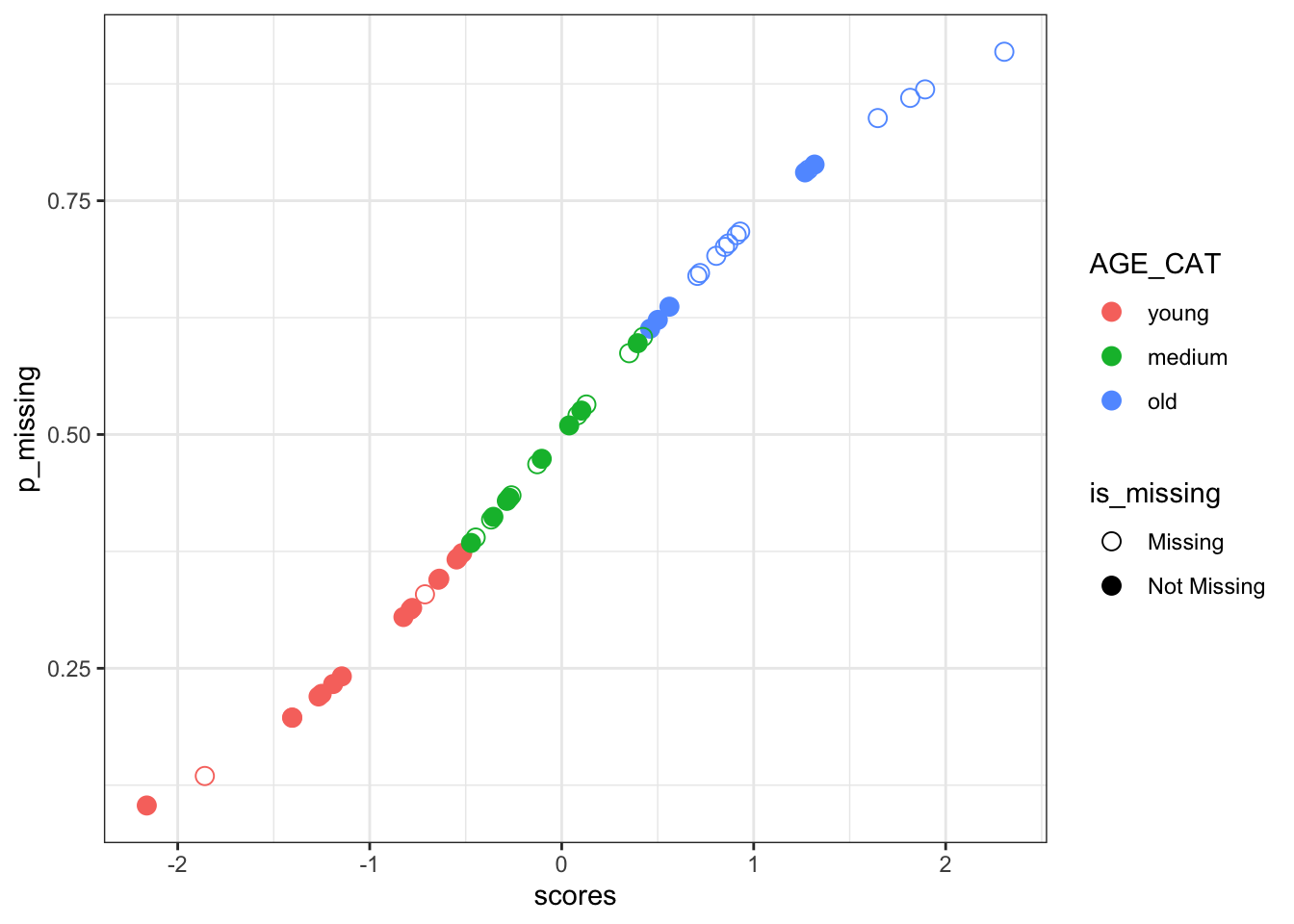
p_missing is_missing
1 0.3446797 0
2 0.4290410 1
3 0.8383969 1
4 0.5097477 0
5 0.5255979 0
6 0.8599921 1mice::ampute and specifically the inner mice::ampute.continuous function goes over a very similar approach but it deals with the so called shifted probability distributions to try to solve the following two issues:
Running the above random generation for the binomial distribution 100 times, we are obtaining a mean proportion of missingness of 51 % (centered at P = 0.5). Then, how does mice::ampute achieve the proportion of missingness assigned by the user? For instance, our target proportion of missingness were 20, 40, 60 and 80%. We need to get the correct shift \(\delta\) parameter so that we can move the probability curves in the y axis.
\[P(missing) = \frac{e^{x + \delta}}{1 + e^{x + \delta}}\]
This can be achieved by the binary search algorithm, and this is how mice::ampute behaves. For the target missing proportion of 60%, the \(\delta\) obtained with the binary search algorithm is of 0.492. Let’s now plot the non-shifted and the shifted probability distributions in the same figure.
shift <- 0.492
p_missing_shift <- plogis(scores + shift)
p_missing_scores_shift <- data.frame("p_missing" = p_missing_shift, "scaled_wss" = scores) %>%
bind_cols(AGE_CAT = amp_df_bind[51:100, c("AGE_CAT", "is_missing")]) %>%
mutate(type = "shifted curve")
p_missing_all <- p_missing_scores_shift %>%
bind_rows(p_missing_scores %>%
mutate(type = "non-shifted curve"))
ggplot(
p_missing_all,
aes(x = scaled_wss, y = p_missing, color = type, group = type)
) +
geom_line() +
theme_bw() +
xlab("Scaled Weighted Sum Scores") + # for the x axis label
ylab("Probability of converting to Missing") 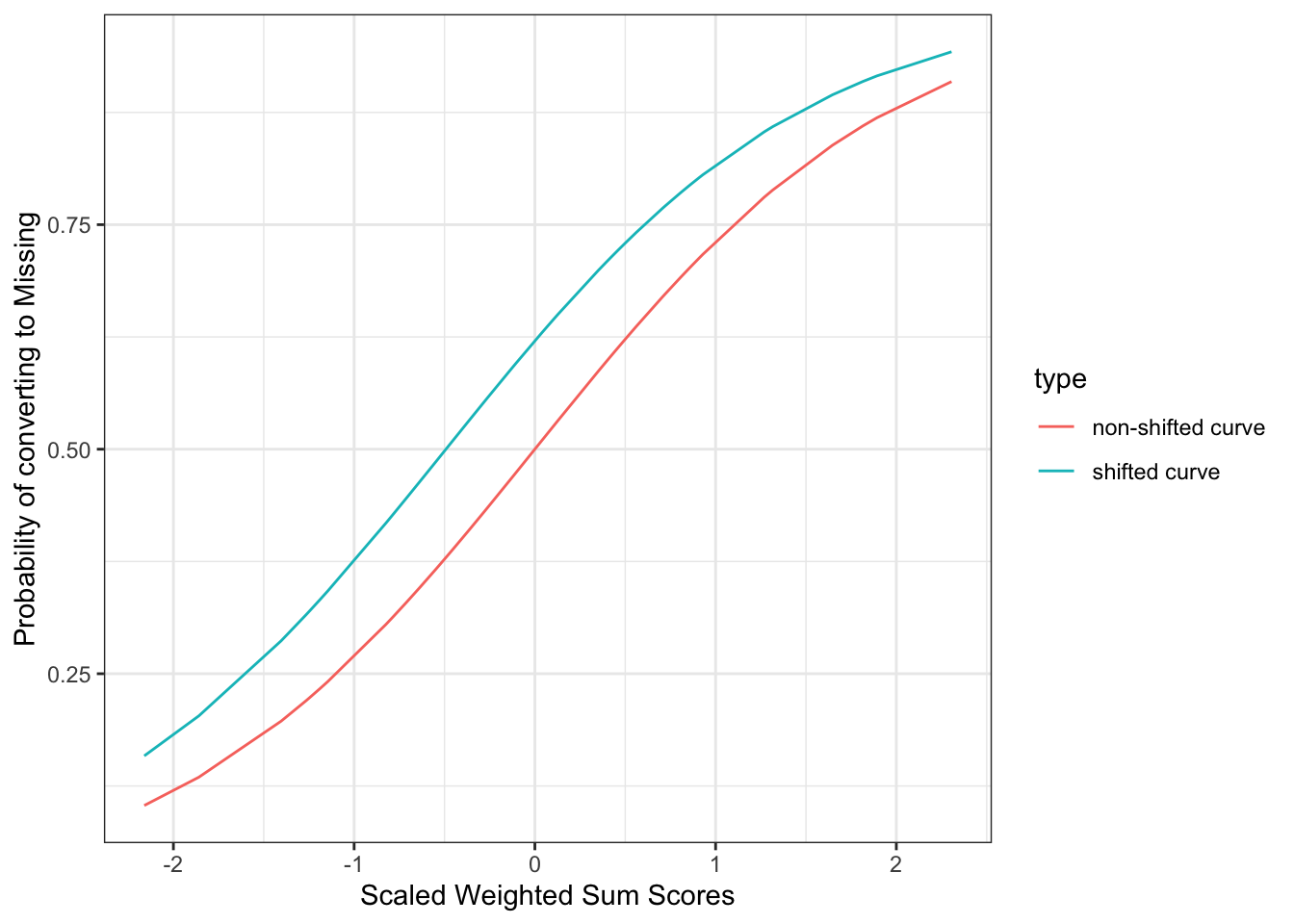
Finally, going now over the random generation process from binomial distribution 100 times, we now successfully achieve a mean value of 60% of missingness! 😎
See the main points we have worked on during this post:
Sharing a picture that I took this weekend at Uetliberg (Zurich, Switzerland). Nice walk and views of the Swiss Alps that helped me putting some of my ideas in order to properly finalize this writing. 😃

See you in the next post!
IZ